A collection of tools for exposome data analysis
Cardiovascular diseases remain the leading cause of death in Europe, representing 45% of all deaths on the continent1. Equally, metabolic diseases – such as type 2 diabetes and obesity – are prevalent, with 61 million people in Europe diagnosed with diabetes2.
The LongITools project is investigating the links between environmental, lifestyle and biological factors (the exposome) and the risk of developing these chronic diseases. Having a better understanding of all of this, could help reduce the strain on healthcare budgets and services across Europe, through the development of targeted policies and interventions to reduce or prevent cardiometabolic diseases.
The LongITools team has developed several tools and methodologies to analyse multiple exposome variables within the data sets being used in the project. We have also developed the Exposome Data Analysis Toolbox, a tool which provides LongITools researchers with access to several data analysis applications, in a single open-source platform. It also allows other researchers and data scientists to benefit from the tools in further studies and projects beyond the LongITools project.
How we developed the toolbox
The Exposome Data Analysis Toolbox was created by the University of Barcelona using existing software components, adapted to suit the needs of the LongITools project. The foundation was provided by an evolved version of the EuCanSHare platform, originally developed for multi-centre cardiovascular data analysis, repurposed for the study.
In turn, the template for how LongITools would function was built on the OpenEBench platform, an open source ‘workbench’ specifically designed to allow for the creation of customised analysis infrastructure. It can quickly create an interactive, web-based virtual research environment, pulling data from local and remote locations for visualisation applications and analysis tools. This ‘one-stop-shop’ approach allows these applications to access data in one click – increasing the usability of the Exposome Data Analysis Toolbox.
The goal of the Toolbox was to create a base from which other tools developed by the project could be attached and integrated, anchoring them to a central user interface. There were several capabilities required from the toolbox. These included:
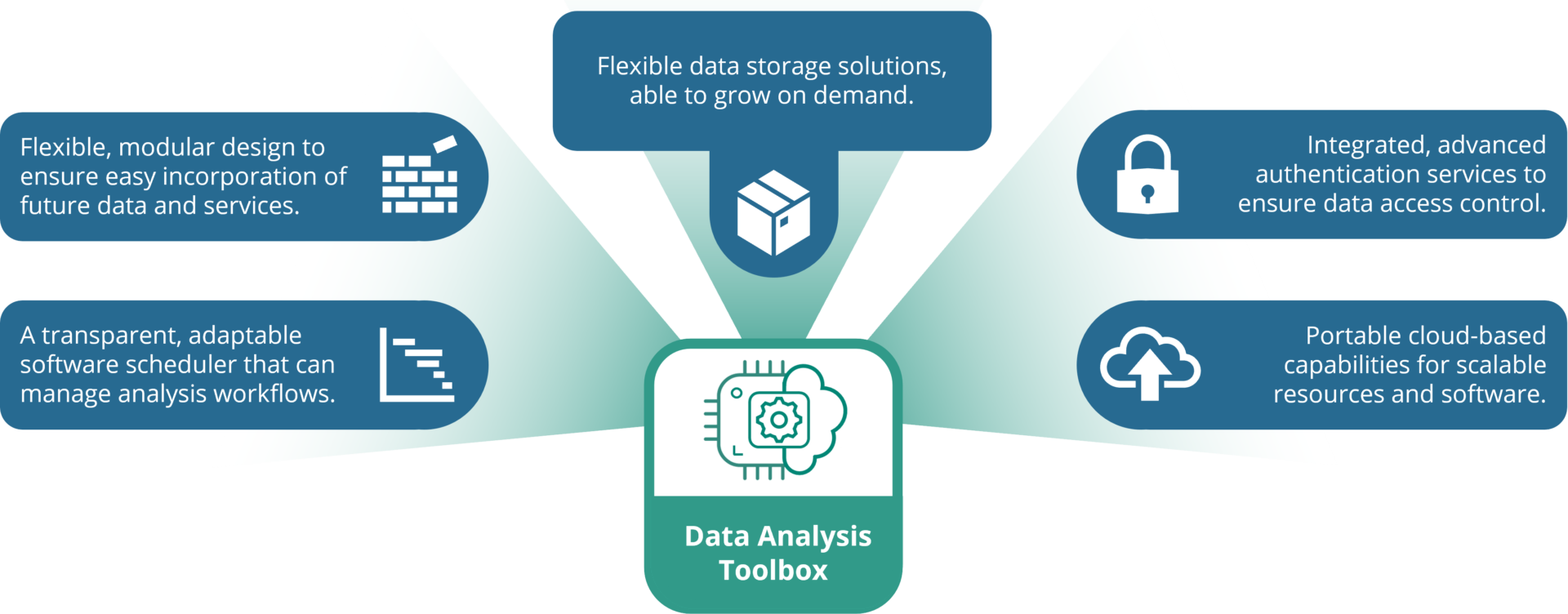
Figure 1: The key features of the LongITools Exposome Data Analysis Toolbox
All these features were aligned with the central objective of the Toolbox – to give exposome researchers access to the tools and methodologies they need, whilst maintaining a focus on data security and confidentiality.
Features of the Toolbox
The Toolbox provides a fully operational, centralised, user-friendly data analysis system. It includes access to tools such as causal inference, machine learning, hierarchical modelling, data visualisation and summary measures which are developed within interfaces such as R/R-Studio and DataSHIELD.
Users can search and access tools and methodologies through the platform, with guidelines, videos and example data to guide them through the process. Whilst the Toolbox is open access, it does require registration to enable some functions, like running a tool during a private session.
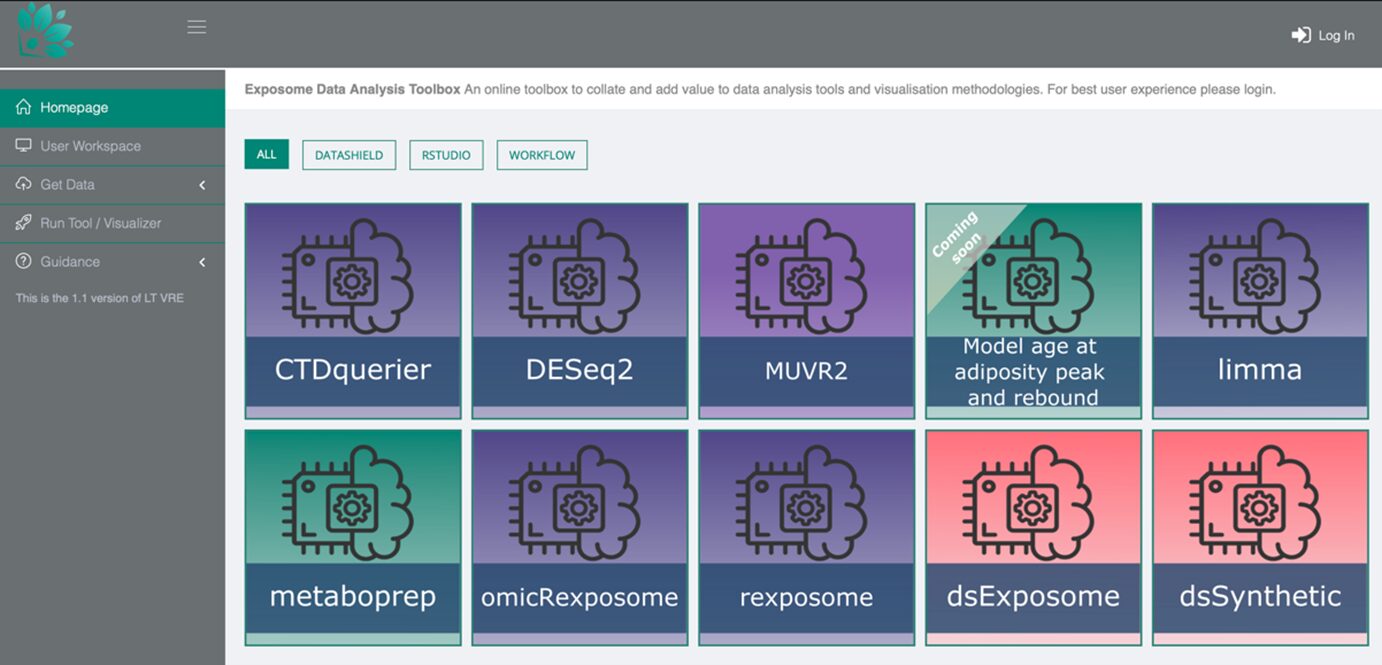
Figure 2: A screenshot of the Toolbox homepage
Currently, the Toolbox contains ten individual applications, with plans to expand with the addition of new data analysis capabilities. These tools currently include:
DataSHIELD Libraries:
- dsExposome helps manage and analyse exposome data, allowing researchers to integrate, pre-process, and analyse a range of environmental exposure data and the health impacts.
- dsSynthetic creates ‘synthetic’ datasets to mimic the real-world exposome data, helping researchers test and validate data analysis, without using sensitive medical information.
R Libraries:
- rexposome contains a library of functions for exposome research – including data cleaning, exposure assessment, statistical analysis and visualisation.
- omicRexposome allows researchers to integrate ‘omics’ data with exposome data, allowing them to analyse their impacts (‘omics includes several areas of research, such as genomics, or metabolomics).
- CTDquerier allows researchers to search the ‘Comparative Toxicogenomic Database’3 for interactions between chemicals, diseases and genes. This will help them to understand and link their exposome data with potential health effects or outcomes.
- The DESeq2 model of discrete gene expression utilises ‘discrete’ gene expression data. This enables the detection of changes in RNA information, particularly in relation to environmental exposures.
- The Limma model of continuous gene expression uses ‘limma’ package software to analyse ‘continuous gene expression’ data, identifying expressed genes and how gene regulation responds to different environmental exposures.
- MUVR2 provides machine learning capabilities and ‘multivariate analysis’ (the examination of multiple factors) when analysing exposome data – supporting predictive modelling and variable selection to help researchers identify key environmental factors that are influencing health outcomes.
Workflows:
- Metaboprep is a pre-processing tool designed for metabolomics data (the study of small molecules within cells). This standardises the data, allowing for clearer statistical analysis.
- A model to show the age at adiposity peak and rebound, predicting the age at which an infant’s BMI reaches its highest point (peak) and when a child’s BMI reaches its lowest point before starting to increase again (rebound). This helps explore the developmental origins of health and disease.
The LongITools Exposome Data Analysis Toolbox provides a flexible, secure and scalable environment for data analysis, helping encourage further collaboration and innovation in exposome research. It offers another step forward in the development of tools to support exposome research – collating several useful tools on one coherent platform that can address the entire data analysis pipeline. It will help create robust, reproducible studies to ultimately better our understanding of the interaction between the exposome and our health.
The University of Barcelona regularly update the toolbox and add new tools with the aim of building a community of researchers who can use and contribute to the growing set of tools for exposome studies. This ensures researchers have access to the most recent and complete set of exposome research tools.
How can I access the Toolbox?
The Toolbox can be accessed via this link: https://longitools.bsc.es/vre/home/
For a short video explainer on how it works, see this video on YouTube: https://youtu.be/uoe5lnr6FQg?si=FNtRdSP26p076Kxi
For a Toolbox summary on the LongITools website, follow this link: https://longitools.org/news/results/exposome-data-analysis-toolbox/
If you wish to include a tool or methodology within the Toolbox please contact: Noussair Lazrak or Karim Lekadir
References
1European Heart Network: https://ehnheart.org/about-cvd/the-burden-of-cvd/
2International Diabetes Foundation: https://diabetesatlas.org/atlas/tenth-edition/
3Comparative Toxicogenomic Database, funded by the NIEHS: https://ctdbase.org/