Simplifying access to multiple research data sets
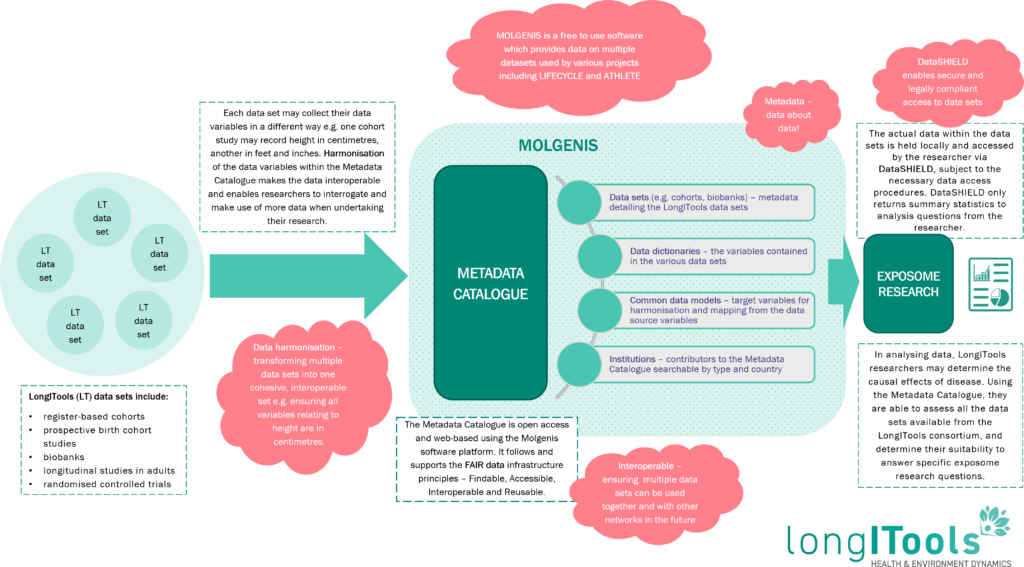
The LongITools consortium has access to a large resource of life-course data, including prospective birth cohort studies and longitudinal studies in adults, register-based cohorts, randomised controlled trials and biobanks. These LongITools data sets include data from 24 different studies on over 11 million EU citizens and a wide range of data variables such as height, weight, blood composition, employment, lifestyle factors, cholesterol and many more.
One of the key objectives of the LongITools project is to integrate and enrich a catalogue of longitudinal multi-dimensional data sets in a findable, accessible, interoperable, and reusable (FAIR) data infrastructure. This infrastructure will reduce the risk of duplication, enable rapid and efficient data discovery, data access and meta-analysis, and intensify collaboration across longitudinal cohorts in Europe.
To achieve this key objective and enable the LongITools exposome researchers to assess all their data sets, and the suitability of the data sets to answer specific research questions, we created a metadata catalogue.
Metadata Catalogue Functions and Features
Rather than developing a separate LongITools catalogue, the team has built on the existing LifeCycle Project’s EU Child Cohort Variable catalogue. In collaboration with ATHLETE (another European Human Exposome Network (EHEN) project) and EUCAN-connect project this existing catalogue was then completely rebuilt. Based on the needs of LongITools and the lessons learned in LifeCycle, the ability to host multiple project consortia within one catalogue was added while refining the underlying data structure. Rich metadata was then added to the catalogue and each data set mapped their exposome variables to an agreed LongITools definition (harmonised model or ‘common’ data model).
The catalogue comprises:
- a findability function enabling users to access rich metadata about the data sets including, for example, the type of data set (e.g., cohort), the population, number of participants, and the data variables;
- a harmonisation mapping system that enables cataloguing on the ‘variable level’, documenting harmonised ‘standard’ variables and providing interoperability maps showing if and how collected variables from the data sets have been mapped to these standards;
- manuals to explain to users how to use the catalogue;
- standard operating procedures.
The catalogue only contains metadata (in simple terms data about data). The actual data within the data sets is held locally and accessed by the researcher independently of the metadata catalogue.
The web-based catalogue is open access, and the catalogue software is built into the MOLGENIS FAIR data platform. Links to the Metadata Catalogue will be available following its public launch.
Metadata Catalogue Explained
For a larger view of this image click here.
FAIR data for a healthier Europe
The FAIR data model used in LongITools has considered existing structures and ensures open access to the data generated. This provides an excellent opportunity to interact and collaborate with the other projects in EHEN, potentially building one unified FAIR human exposome data toolbox that can be used by researchers and policymakers in their actions to make Europe healthier.
So, what does using a FAIR data model actually mean in the LongITools project?
FAIR data in LongITools

Sustainable, Collaborative Tool
By bringing together all existing networks and cohorts, in one FAIR data infrastructure, it will ensure better data reuse and increased benefit to the scientific community. We hope that our approach to create a sustainable catalogue, usable beyond LongITools, will inspire more consortia to participate. The mission is to further accelerate multi-centre exposome and health research in Europe by improving the FAIRness of all rich data available, starting with other EHEN projects.
If you are interested in collaborating with us or learning more please submit an enquiry form via the LongITools Innovation Toolbox.
Find out more about the LongITools data sets.